Biological Effects of Clustered DNA Damage Induced by Space Radiation – Publicly Invited Research 2016-2017
- A02 Shinohara
- A02 Maekawa
- A02 Ohgami
- A02 Nishimura
- A02 Kawano
- A02 Iwase
- A02 Furuichi
- A02 Myung
- A02 Kitamura
- A03 Nakamura
- A03 Harada
- A03 Ide
- A03 Shirai
- A03 Kakinuma
| Research Subject | Biological Effects of Clustered DNA Damage Induced by Space Radiation |
|---|---|
| Research Group Leader |

|
| Research Collaborator(s) |
|
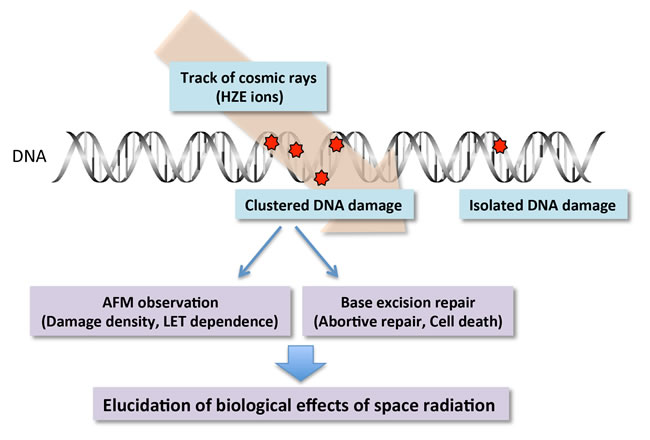
The health effect of cosmic rays is a major concern to astronauts on an interplanetary mission or a long-term mission to the International Space Station. Galactic cosmic rays (GCRs) consist of high-energy protons, helium nuclei, and high atomic number (Z) and energy ions (HZE ions). Although HZE ions make up a small fraction of GCRs, they would contribute significantly to the overall biological impact of cosmic rays due to their high Relative Biological Effectiveness (RBE). HZE ions are high linear energy transfer (LET) radiations and cause dense ionization along their track, resulting in the formation of clustered DNA damage. The site of clustered DNA damage contains two or more lesions within a few helical turns of the DNA after passage of a single radiation track. They are signatures of DNA modifications induced by ionizing radiation (particularly by HZE ions).
The structural complexity, repairability, and biological consequences of clustered DNA damage would vary depending on the type of HZE ions and their LET. However, very limited information is available on these subjects since there is no experimental method to analyze the extent of the structural complexity of clustered DNA damage. A certain type of clustered DNA damage can be detected as a DNA double-strand break (DSB) upon treatment with repair enzyme. However, this does not provide any information about the structural complexity.
In the present study we will address two issues. First, to gain insight into the structural complexity of clustered DNA damage generated by HZE ions, individual DNA lesions in a clustered DNA damage site are specifically labeled with biotin/avidin and directly observed by atomic force microscopy (AFM). The structural complexity is assessed on the basis of the number of DNA lesions per clustered DNA damage site. Second, we will clarify whether simultaneous processing of two or more lesions in a clustered DNA damage site by base excision repair (BER) results in a DSB that is more toxic to cells than original lesions (abortive repair). Cells proficient and deficient in BER are irradiated with HZE ions and the effect of abortive repair is elucidated on the basis of cell survival and genome damage.